Preprints
|
| P11. |
AniAnn's: alignment-free annotation of tandem repeat arrays using fast average nucleotide identity estimates
Sweeten, A, Schatz, MC, Phillippy AM (2026) bioRxiv doi:10.64898/2026.01.27.702063 |
 |
|
| P10. |
Behavioral Assessment Reliability in Clinical Phenotyping and Biomarker Research for Autism
Perez-Benavides, E et al (2026) medRxiv doi: https://doi.org/10.64898/2026.01.24.25343227 |
 |
|
| P9. |
Standardizing RNA-seq Analysis of Fungal Pathogens Using BRC-Analytics and Agentic AI: A Candidozyma auris Case Study
Nekrutenko et al (2025) bioRxiv doi: 10.64898/2025.12.30.697050 |
 |
|
| P8. |
Genomic Next-Token Predictors are In-Context Learners
Breslow et al (2025) arXiv doi: https://arxiv.org/abs/2511.12797 |
 |
|
| P7. |
Insights into the Datasets, Tools, and Training Needs of the AnVIL Community: 2024
Isaac et al (2025) bioRxiv doi: https://doi.org/10.1101/2025.11.14.688517 |
 |
|
| P6. |
From data to publication in a browser with BRC-Analytics: Evolutionary dynamics of coding overlaps in measles virus
Nekrutenko et al (2025) bioRxiv doi: https://doi.org/10.1101/2025.10.13.682095 |
 |
|
| P5. |
Population-scale Long-read Sequencing in the All of Us Research Program
Garimella, Li, Wertz, Lee, Cunial et al (2025) bioRxiv doi:10.1101/2025.09.21.677443 |
 |
|
| P4. |
A complete diploid human genome benchmark for personalized genomics
Hansen et al (2025) bioRxiv doi:10.1101/2025.09.21.677443 |
 |
|
| P3. |
Assembling unmapped reads reveals hidden variation in South Asian genomes
Das, A, Biddanda, A, McCoy, RC, Schatz, MC (2025) bioRxiv doi: https://doi.org/10.1101/2025.05.14.653340 |
 |
|
| P2. |
microGalaxy: A gateway to tools, workflows, and training for reproducible and FAIR analysis of microbial data
Nasr et al (2024) bioRxiv https://doi.org/10.1101/2024.12.23.629682 |
 |
|
| P1. |
The genome of the Wollemi pine, a critically endangered living fossil unchanged since the Cretaceous, reveals extensive ancient transposon activity
Stevenson, DW, Ramakrishnan, S, de Santis Aleves, C et al.. (2023) bioRxiv doi: https://doi.org/10.1101/2023.08.24.554647 |
 |
|
|
|
2025
|
| 201. |
Unearthing soil biodiversity through collaborative genomic research and education
The BioDIGS Consortium (2025) Nature Genetics https://doi.org/10.1038/s41588-025-02442-5 |
 |
|
| 200. |
Engineering compact Physalis peruviana (goldenberry) to promote its potential as a global crop
Santo Domingo et al (2025) Plants, People, Planet https://doi.org/10.1002/ppp3.70140 |
 |
|
| 199. |
Learning Explainable Imaging-Genetics Associations Related to a Neurological Disorder
Wang et al (2025) MICCAI https://doi.org/10.1007/978-3-032-05141-7_34 |
 |
|
| 198. |
Comparative genomics of the parasite Trichomonas vaginalis reveals genes involved in spillover from birds to humans
Sullivan et al (2025) Nature Communications https://doi.org/10.1038/s41467-025-61483-w |
 |
|
| 197. |
Complete sequencing of ape genomes
Yoo et al (2025) Nature doi: 10.1038/s41586-025-08816-3 |
 |
|
| 196. |
Uncalled4 improves nanopore DNA and RNA modification detection via fast and accurate signal alignment
Kovaka, S, Hook, PW, Jenike, KM, Shivakumar, V, Morina, LB, Razahhi, R, Timp, W, Schatz, MC (2025) Nature Methods https://doi.org/10.1038/s41592-025-02631-4 |
 |
|
| 195. |
Integration of transcriptomics and long-read genomics prioritizes structural variants in rare disease
Jensen, Ni, et al. (2025) Genome Research doi: 10.1101/gr.279323.124 |
 |
|
| 194. |
Unraveling the hidden complexity of cancer through long-read sequencing
Li et al (2025) Genome Research doi: 10.1101/gr.280041.124
|
 |
|
| 193. |
The Farm Animal Genotype–Tissue Expression (FarmGTEx) Project
Fang et al (2025) Nature Genetics https://doi.org/10.1038/s41588-025-02121-5
|
 |
|
| 192. |
Solanum pan-genetics reveals paralogues as contingencies in crop engineering
Benoit, Jenike et al (2025) Nature https://doi.org/10.1038/s41586-025-08619-6
Nature News & Views: Genus-wide plant pangenome could inform next-generation crop design |
 |
|
| 191. |
k-mer approaches for biodiversity genomics
Jenike et al. (2024) Genome Research doi: 10.1101/gr.279452.124 |
 |
|
|
|
2024
|
| 190. |
MaizeCODE reveals bi-directionally expressed enhancers that harbor molecular signatures of maize domestication
Cahn et al. (2024) Nature Communication https://doi.org/10.1038/s41467-024-55195-w |
 |
|
| 189. |
Gapless assembly of complete human and plant chromosomes using only nanopore sequencing
Koren et al. (2024) Genome Research doi: 10.1101/gr.279334.124 |
 |
|
| 188. |
BEATRICE: Bayesian Fine-mapping from Summary Data using Deep Variational Inference
Ghosal, S, Schatz, M, Venkataraman, A (2024) Bioinformatics https://doi.org/10.1093/bioinformatics/btae590 |
 |
|
| 187. |
High-coverage nanopore sequencing of samples from the 1000 Genomes Project to build a comprehensive catalog of human genetic variation
Gustafson et al. (2024) Genome Research doi:10.1101/gr.279273.124 |
 |
|
| 186. |
MEM-based pangenome indexing for k-mer queries
Hwang et al (2024) International Workshop on Algorithms in Bioinformatics (WABI) https://doi.org/10.4230/LIPIcs.WABI.2024.4
Journal version in Algorithms for Molecular Biology |
 |
|
| 185. |
Differences in activity and stability drive transposable element variation in tropical and temperate maize.
Ou, S, Collins, T, Qiu, Y, Seetharam, AS, Menard, CC, Manchanda, N, Gent, JI, Schatz, MC, Anderson, SN, Hufford, MB, Hirsch CN (2024) Genome Research doi: 10.1101/gr.278131.123 |
 |
|
| 184. |
ModDotPlot - Rapid and interactive visualization of tandem repeats
Sweeten, AP, Schatz, MC, Phillippy, AM (2024) Bioinformatics doi:10.1093/bioinformatics/btae493 |
 |
|
| 183. |
Convergent evolution of plant prickles by repeated gene co-option over deep time
Satterlee et al. (2024) Science doi: 10.1126/science.ado1663 |
 |
|
| 182. |
Interspecies regulatory landscapes and elements revealed by novel joint systematic integration of human and mouse blood cell epigenomes
Xiang et al. (2024) Genome Research doi:10.1101/gr.277950.123 |
 |
|
| 181. |
The complete sequence and comparative analysis of ape sex chromosomes
Mokova et al. (2024) Nature doi: 10.1038/s41586-024-07473-2 |
 |
|
| 180. |
The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update
The Galaxy Community (2024) Nucleic Acids Research https://doi.org/10.1093/nar/gkae410 |
 |
|
| 179. |
Beyond the Human Genome Project: The Age of Complete Human Genome Sequences and Pangenome References
Taylor, DJ, Eizenga, JM, Li, Q, et al. (2024) Annual Review of Genomics and Human Genetics doi: 10.1146/annurev-genom-021623-081639 [reprint] |
 |
|
| 178. |
Utility of long-read sequencing for All of Us
Mahmoud et al. (2024) Nature Communication doi:10.1038/s41467-024-44804-3 |
 |
|
| 177. |
Scalable, accessible, and reproducible reference genome assembly and evaluation in Galaxy
Lariviere et al. (2024) Nature Biotechnology doi:10.1038/s41587-023-02100-3 |
 |
|
|
|
2023
|
| 176. |
Long-read sequencing reveals rapid evolution of immunity- and cancer-related genes in bats
Scheben, A, Ramos, OM, Kramer, M, Goodwin, S, Oppenheim, S, Becker, DJ, Schatz, MC, Simmons, NB, Siepel, A, McCombie, WR (2023) Genome Biology and Evolution doi: 10.1093/gbe/evad148 |
 |
|
| 175. |
The complete sequence of a human Y chromosome
Rhie, A*, Nurk, S*, Cechova, M*, Hoyt, SJ*, Taylor, DJ*, et al. (2023) Nature https://doi.org/10.1038/s41586-023-06457-y |
 |
|
| 174. |
Concerning the eXclusion in human genomics: The choice of sex chromosome representation in the human genome drastically affects number of identified variants
Pinto, BJ, O'Connor, B, Schatz, M, Zarate, S, Wilson, MW. (2023) bioRxiv https://doi.org/10.1101/2023.02.22.529542 |
 |
|
| 173. |
Characterization of large-scale genomic differences in the first complete human genome
Yang et al. (2023) Genome Biology https://doi.org/10.1186/s13059-023-02995-w |
 |
|
| 172. |
Fast and accurate genome-wide predictions and structural modeling of protein-protein interactions using Galaxy
Guerler, A, Baker, D, van den Beek, M, Bouvier, D, Coraor, N, Schatz, MC, Nekrutenko, A (2023) BMC Bioinformatics doi: https://doi.org/10.1186/s12859-023-05389-8 |
 |
|
| 171. |
Genome analyses reveal population structure and a purple stigma color gene candidate in finger millet
Davos, KM, Qi, P, Bahri, BA et al. (2023) Nature Communication doi: 10.1038/s41467-023-38915-6 |
 |
|
| 170. |
Multicentre genetic diversity study of carbapenem-resistant Enterobacterales: predominance of untypeable pUVA-like blaKPC bearing plasmids
Simner PJ, Bergman Y, Fan Y, Jacobs EB, Ramakrishnan S, Lu J, Lewis S, Hanlon A, Tamma PD, Schatz MC, Timp W, Carroll KC (2023) JAC Antimicrobial Resistance doi: 10.1093/jacamr/dlad061 |
 |
|
| 169. |
NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-Space: Reaching out to Clinicians
Hall, JL, Honeycutt, S, Gonzalez, N, O’Donnell-Luria, A, Overby Taylor, C, Stevens, L, Philippakis, AA, Schatz, MC (2023) Circulation: Genomic and Precision Medicine https://doi.org/10.1161/CIRCGEN.122.003936 |
 |
|
| 168. |
The EN-TEx resource of multi-tissue personal epigenomes & variant-impact models
Rozowsky et al (2023) Cell doi:https://doi.org/10.1016/j.cell.2023.02.018 |
 |
|
| 167. |
Jasmine and Iris: Population-scale structural variant comparison and analysis
Kirsche, M, Prabhu, G, Sherman, R, Ni, B, Battle, A, Aganezov, S, Schatz, MC (2023) Nature Methods https://doi.org/10.1038/s41592-022-01753-3 |
 |
|
| 166. |
Approaching complete genomes, transcriptomes and epi-omes with accurate long-read sequencing
Kovaka, S, Ou, S, Jenike, K,Schatz, MC (2023) Nature Methods https://doi.org/10.1038/s41592-022-01716-8 |
 |
|
|
|
2022
|
| 165. |
Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing
Alonge, M, Lebeigle, L, Kirsche, M, Jenike, K, Ou, S, Aganezov, S, Wang, X, Lippman, ZB, Schatz, MC+, Soyk, S+ (2022) Genome Biology doi: https://doi.org/10.1186/s13059-022-02823-7 |
 |
|
| 164. |
Sketching and sampling approaches for fast and accurate long read classification
Das, A, Schatz, MC (2022) BMC Bioinformatics doi: https://doi.org/10.1186/s12859-022-05014-0 |
 |
|
| 163. |
Establishing Physalis as a new Solanaceae model system enables genetic reevaluation of the inflated calyx syndrome
He, J, Alonge, M, Ramakrishnan, S, Benoit, M, Soyk, S, Reem, N, Hendelman, A, Van Eck, J, Schatz, M, Lippman, Z (2022) The Plant Cell https://doi.org/10.1093/plcell/koac305 |
 |
|
| 162. |
Semi-automated assembly of high-quality diploid human reference genomes
Jarvis, ED et al. (2022) Nature https://doi.org/10.1038/s41586-022-05325-5 |
 |
|
| 161. |
Diversifying the Genomic Data Science Research Community
The Genomics Data Science Community Network (2022) Genome Research doi: 10.1101/gr.276496.121 |
 |
|
| 160. |
Minos: variant adjudication and joint genotyping of cohorts of bacterial genomes
Hunt, M, Letcher, B, Malone, KM, Nguyen, G, Hall, MB, Colquhoun, RM, Lima, L, Schatz, MC, Ramakrishnan, S, CRyPTIC consortium, Iqbal, Z (2022) Genome Biology doi: https://doi.org/10.1186/s13059-022-02714-x |
 |
|
| 159. |
Benchmarking challenging small variants with linked and long reads
Wagner et al (2022) Cell Genomics doi: https://doi.org/10.1016/j.xgen.2022.100128 |
 |
|
| 158. |
The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update
The Galaxy Community (2022) Nucleic Acids Research https://doi.org/10.1093/nar/gkac247 |
 |
|
| 157. |
Complete sequence of a 641-kb insertion of mitochondrial DNA in the Arabidopsis thaliana nuclear genome
Fields, PD, Waneka, G, Naish, M, Schatz, MC, Henderson, IR, Sloan, DB (2022) Genome Biology and Evolution https://doi.org/10.1093/gbe/evac059 |
 |
|
| 156. |
The complete sequence of a human genome
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM (2022) Science doi: 10.1126/science.abj6987 |
 |
|
| 155. |
A complete reference genome improves analysis of human genetic variation
Aganezov, S*, Yan, SM*, Soto, DC*, Kirsche, M*, Zarate, S*, Avdeyev, P, Taylor, DJ, Shafin, K, Shumate, A, Xiao, C, Wagner, J, McDaniel, J, Olson, ND, Sauria, MEG, Vollger, MR, Rhie, A, Meredith, M, Martin, S, Lee, J, Koren, S, Rosenfeld, J, Paten, B, Layer, R, Chin, CS, Sedlazeck, FJ, Hansen, NF, Miller, DE, Phillippy, AM, Miga, KM, McCoy, RC†, Dennis, MY†, Zook, JW†, Schatz, MC† (2022) Science doi: 10.1126/science.abl3533 |
 |
|
| 154. |
Epigenetic Patterns in a Complete Human Genome
Gershman, A, Sauria, MEG, Hook, PW, Hoyt, S, Razaghi, R, Koren, S, Altemose, N, Valadas, GV, Vollger, MR, Logsdon, GA, Rhie, A, Eichler, EE, Schatz, MC, O'Neill, RJ, Phillippy, AM, Miga, KH, Timp, W (2022) Science doi: 10.1126/science.abj5089 |
 |
|
| 153. |
Complete genomic and epigenetic maps of human centromeres
Altemose N, Logsdon GA, Bzikadze AV, Sidhwani P, Langley SA, Caldas GV, Hoyt SJ, Uralsky L, Ryabov FD, Shew CJ, Sauria MEG, Borchers M, Gershman A, Mikheenko A, Shepelev VA, Dvorkina T, Kunyavskaya O, Vollger MR, Rhie A, McCartney AM, Asri M, Lorig-Roach R, Shafin K, Lucas JK, Aganezov S, Olson D, de Lima LG, Potapova T, Hartley GA, Haukness M, Kerpedjiev P, Gusev F, Tigyi K, Brooks S, Young A, Nurk S, Koren S, Salama SR, Paten B, Rogaev EI, Streets A, Karpen GH, Dernburg AF, Sullivan BA, Straight AF, Wheeler TJ, Gerton JL, Eichler EE, Phillippy AM, Timp W, Dennis MY, O'Neill RJ, Zook JM, Schatz MC, Pevzner PA, Diekhans M, Langley CH, Alexandrov IA, Miga KH (2022) Science doi: 10.1126/science.abl4178 |
 |
|
| 152. |
From telomere to telomere: the transcriptional and epigenetic state of human repeat elements
Hoyt SJ, Storer JM, Hartley GA, Grady PGS, Gershman A, de Lima LG, Limouse C, Halabian R, Wojenski L, Rodriguez M, Altemose N, Rhie A, Core LJ, Gerton JL, Makalowski W, Olson D, Rosen J, Smit AFA, Straight AF, Vollger MR, Wheeler TJ, Schatz MC, Eichler EE, Phillippy AM, Timp W, Miga KH, O'Neill RJ (2022) Science doi: 10.1126/science.abk3112 |
 |
|
| 151. |
Same-Cell Co-Occurrence of RAS Hotspot and BRAF V600E Mutations in Treatment-Naive Colorectal Cancer
Gularte-Mérida R, Smith S, Bowman AS, da Cruz Paula A, Chatila W, Bielski CM, Vyas M, Borsu L, Zehir A, Martelotto LG, Shia J, Yaeger R, Fang F, Gardner R, Luo R, Schatz MC, Shen R, Weigelt B, Sánchez-Vega F, Reis-Filho JS, Hechtman JF. (2022) JCO Precis Oncol. doi:10.1200/po.21.00365 |
 |
|
| 150. |
Artificial Intelligence and Cardiovascular Genetics
Krittanawong, C, Johnson, KW, Choi, E, Kaplin, S, Venner, E, Murugan, M, Wang, Z, Glicksberg, BS, Amos, CI, Schatz, MC, Tang, W (2022) Life https://doi.org/10.3390/life12020279 |
 |
|
| 149. |
Inverting the model of genomics data sharing with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space (AnVIL)
Schatz, MC*, Philippakis, AA*, Afgan, A, Banks, E, Carey, VJ, Carroll, RJ, Culotti, A, Ellrott, K, Goecks, J, Grossman, RL, Hall, IM, Hansen, KD, Lawson, J, Leek, JT, O’Donnell Luria, A, Mosher, S, Morgan, M, Nekrutenko, A, O’Connor, BD, Osborn, K, Paten, B, Patterson, C, Tan, FJ, Taylor, CO, Vessio, J, Waldron, L, Wang, T, Wuichet, K, AnVIL Team (2022) Cell Genomics doi: https://doi.org/10.1016/j.xgen.2021.100085 |
 |
|
|
|
2021
|
| 148. |
The genetic and epigenetic landscape of the Arabidopsis centromeres
Naish, M*, Alonge, M*, Wlodzimierz, P*, Tock, AJ, Abramson, BW, Lambing, CA, Kuo, P, Yelina, N, Hartwick, N, Colt, K, Kakutani, T, Martienssen, RA, Bousios, A, Michael, TP, Schatz, MC+, Henderson, IR+ (2021) Science doi: 10.1126/science.abi7489 |
 |
|
| 147. |
Democratizing long-read genome assembly
Kirsche, M, Schatz, MC (2021) Cell Systems https://doi.org/10.1016/j.cels.2021.09.010 |
 |
|
| 146. |
High resolution copy number inference in cancer using short-molecule nanopore sequencing
Baslan, T, Kovaka, S, Sedlazeck, FJ, Zhang, Y, Wappel, R, Lowe, SW, Goodwin, S, Schatz, MC (2021) Nucleic Acids Research https://doi.org/10.1093/nar/gkab812 |
 |
|
| 145. |
Local adaptation and archaic introgression shape global diversity at human structural variant loci
Yan, S, Sherman, RS, Taylor, DJ, Nair, DR, Bortvin, AN, Schatz, MC, McCoy, RC (2021) eLife doi:10.7554/eLife.67615 |
 |
|
| 144. |
Pan-genomic Matching Statistics for Targeted Nanopore Sequencing
Ahmed, O, Rossi, M, Kovaka, S, Schatz, M, Gagie, T, Boucher, C, Langmead, B (2021) iScience doi: https://doi.org/10.1016/j.isci.2021.102696 |
 |
|
| 143. |
An anchored chromosome-scale genome assembly of spinach improves annotation and reveals extensive gene rearrangements in euasterids
Hulse-Kemp, AM, Bostan, H, Chen, S, Ashrafi, H, Stoffel, K, Sanseverino, W, Li, L, Cheng, S, Schatz, MC, Garvin, T, du Toit, LJ, Tseng, E, Chin, J, Iorizzo, M, Van Deynze, A (2021) The Plant Genome https://doi.org/10.1002/tpg2.20101 |
 |
|
| 142. |
A plasmid locus associated with Klebsiella clinical infections encodes a microbiome-dependent gut fitness factor
Vornhagen, J, Bassis, CM, Ramakrishnan, S, Hein, R, Mason, S, Bergman, Y, Sunshine, N, Fan, Y, Holmes, CL, Timp, W, Schatz, MC, Young, VB, Simner, PJ, Bachman, MA (2021) PLOS Pathogens https://doi.org/10.1371/journal.ppat.1009537 |
 |
|
| 141. |
The genomic basis of evolutionary differentiation among honey bees
Fouks, B, Brand, P, Nguyen, NN, Herman, J, Camara, F, Ence, D, Hagen, D, Hoff, KJ, Nachweide, S, Romoth, L, Walden, KK, Guigo, R, Stanke, M, Narzisi, G, Yandell, M, Robertson, HM, Koeniger, N, Chantawannakul, P, Schatz, MC, Worley, KC, Robinson, GE, Elsik, CG, Rueppell, O (2021) Genome Research doi: 10.1101/gr.272310.120 |
 |
|
| 140. |
Natural genetic diversity in tomato flavor genes
Pereira, L, Sapkota, M, Alonge, M, Zheng, Y, Zhang, Y, Razifard, H, Taitano, NK, Schatz, M, Fernie, A, Wang, Y, Fei, Z, Caicedo, AL, Tieman, D, Van Der Knaap, E(2021) Front. Plant Sci. doi: 10.3389/fpls.2021.642828 |
 |
|
| 139. |
Optimized sample selection for cost-efficient long-read population sequencing
Ranallo-Benavidez, T, Lemmon, Z, Soyk, S, Aganezov, S, Salerno, WJ, McCoy, RC, Lippman, Z, Schatz, MC, Sedlazeck, FJ (2021) Genome Research doi: 10.1101/gr.264879.120 |
 |
|
| 138. |
Genomic Diversity of SARS-CoV-2 During Early Introduction into the United States National Capital Region
Thielen, PM, Wohl, S, Mehoke, T, Ramakrishnan, S, Kirsche, M, Falade-Nwulia, O, Trovao, NS, Erlund, A, Howser, C, Sadowski, N, Morris, P, Hopkins, M, Schwartz, M, Fan, Y, Gniazdowski, V, Lessler, J, Sauer, L, Schatz, MC, Evans, JD, Ray, SC, Timp, W, Mostafa, HN (2021) JCI Insight doi: 10.1172/jci.insight.144350 |
 |
|
| 137. |
Cell wall protein variation, break‐induced replication, and subtelomere dynamics in Candida glabrata
Xu, X, Green, B, Benoit, N, Sobel, JD, Schatz, MC, Wheelan, S, Cormack, BP (2021) Molecular Microbiology https://doi.org/10.1111/mmi.14707 |
 |
|
| 136. |
SNPC-1.3 is a sex-specific transcription factor that drives male piRNA expression in C. elegans
Choi, CP, Tay, RJ, Starostik, MR, Feng, S, Moresco, JJ, Montgomery, BE, Xu, E, Hammond, MA, Schatz, MC, Montgomery, TA, Yates, JR, Jacobsen, SE, Kim, JK (2021) eLife doi: 10.7554/eLife.60681 |
 |
|
| 135. |
Using Galaxy to Perform Large‐Scale Interactive Data Analyses—An Update
Ostrovsky, A, Hillman‐Jackson, J, Bouvier, D, Clements, D, Afgan, E, Blankenberg, D, Schatz, MC, Nekrutenko, A, Taylor, J, The Galaxy Team, Lariviere, D (2021) Current Protocols https://doi.org/10.1002/cpz1.31 |
 |
|
| 134. |
The human Origin Recognition Complex is essential for pre-RC assembly, mitosis and maintenance of nuclear structure.
Chou, HC, Bhalla, K, El Demerdesh, O, Klingbeil, O, Hanington, K, Aganezov, S, Andrews, P, Alsudani, H, Chang, K, Vakoc, CR, Schatz, MC McCombie, WR, Stillman, B (2021) eLife doi: 10.7554/eLife.61797 |
 |
|
|
|
2020
|
| 133. |
Skyhawk: An Artificial Neural Network-based discriminator for reviewing clinically significant genomic variants
Luo, R, Lam, TW, Schatz, MC (2020) International Journal of Computational Biology and Drug Design doi: 10.1504/IJCBDD.2020.113818 |
 |
|
| 132. |
Parliament2: Accurate structural variant calling at scale
Zarate S, Carroll A, Mahmoud M, Krasheninina O, Jun G, Salerno WJ, Schatz MC, Boerwinkle E, Gibbs RA, Sedlazeck FJ (2020) GigaScience doi: 10.1093/gigascience/giaa145 |
 |
|
| 131. |
iGenomics: Comprehensive DNA Sequence Analysis on your Smartphone
Palatnick, A, Zhou, B, Ghedin, E, Schatz, MC (2020) GigaScience doi: https://doi.org/10.1093/gigascience/giaa138 |
 |
|
| 130. |
Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED
Kovaka, S, Fan, Y, Ni, B, Timp, W, Schatz, MC (2020) Nature Biotechnology doi: https://doi.org/10.1038/s41587-020-0731-9 |
 |
|
| 129. |
Clonal Hematopoiesis Before, During, and After Human Spaceflight
Trinchant et al (2020) Cell Reports doi: https://doi.org/10.1016/j.celrep.2020.108458 |
 |
|
| 128. |
Sapling: Accelerating Suffix Array Queries with Learned Data Models
Kirsche, M, Das, A, Schatz, MC (2020) Bioinformatics doi: https://doi.org/10.1093/bioinformatics/btaa911 |
 |
|
| 127. |
A diploid assembly-based benchmark for variants in the major histocompatibility complex
Chin et al. (2020) Nature Communication doi: https://doi.org/10.1038/s41467-020-18564-9 |
 |
|
| 126. |
Comprehensive analysis of structural variants in breast cancer genomes using single molecule sequencing
Aganezov, S, Goodwin, S, Sherman, R, Sedlazeck, FJ, Arun, G, Bhatia, S, Lee, I, Kirsche, M, Wappel, R, Kramer, M, Kostroff, K, Spector, DL, Timp, W, McCombie, WR, Schatz, MC (2020) Genome Research doi: 10.1101/gr.260497.119 |
 |
|
| 125. |
Ribbon: Intuitive visualization for complex genomic variation
Nattestad, M, Aboukhalil, R, Chin, CS, Schatz, MC (2020) Bioinformatics https://doi.org/10.1093/bioinformatics/btaa680 |
 |
|
| 124. |
Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato
Alonge, M, Wang, X, et al (2020) Cell https://doi.org/10.1016/j.cell.2020.05.021 |
 |
|
| 123. |
A robust benchmark for germline structural variant detection
Zook, J, et al (2020) Nature Biotechnology https://doi.org/10.1038/s41587-020-0538-8 |
 |
|
| 122. |
Vargas: heuristic-free alignment for assessing linear and graph read aligners
Darby, CA, Gaddipati, R, Schatz, MC, Langmead, B (2020) Bioinformatics https://doi.org/10.1093/bioinformatics/btaa265 |
 |
|
| 121. |
In memory of James Taylor: the birth of Galaxy
Nekrutenko, A, Schatz, MC (2020) Genome Biology doi: https://doi.org/10.1186/s13059-020-02016-0 |
 |
|
| 120. |
Management, Analyses, and Distribution of the MaizeCODE Data on the Cloud
Wang et al. (2020) Frontiers of Plant Science doi: https://doi.org/10.3389/fpls.2020.00289 |
 |
|
| 119. |
GenomeScope 2.0 and Smudgeplots: Reference-free profiling of polyploid genomes
Ranallo-Benavidez, TR, Karon, KS, Schatz, MC (2020) Nature Communication doi: https://doi.org/10.1038/s41467-020-14998-3 |
 |
|
| 118. |
De novo genome assembly of Candida glabrata reveals cell wall protein complement and structure of dispersed tandem repeat arrays
Xu, Z, Green, B, Benoit, N, Schatz, M, Wheelan, S, Cormack, B (2020) Molecular Microbiology doi:10.1111/mmi.14488 |
 |
|
|
|
2019
|
| 117. |
Paragraph: A graph-based structural variant genotyper for short-read sequence data
Chen, S, Krusche, P, Dolzhenko, E, Sherman, RM, Petrovski, R, Schlesinger, F, Kirsche, M, Bentley, DR, Schatz, MC, Sedlazeck, FJ, Eberle, MA (2019) Genome Biology doi:10.1186/s13059-019-1909-7 |
 |
|
| 116. |
Recovering rearranged cancer chromosomes from karyotype graphs
Aganezov, S, Zban, I, Aksenov, V, Alexeev, N, Schatz, MC (2019) BMC Bioinformatics doi:10.1186/s12859-019-3208-4 |
 |
|
| 115. |
Assembly of the 373k gene space of the polyploid sugarcane genome reveals reservoirs of functional diversity in the world's leading biomass crop
Souza et al. (2019) GigaScience https://doi.org/10.1093/gigascience/giz129 |
 |
|
| 114. |
RaGOO: Fast and accurate reference-guided scaffolding of draft genomes
Alonge, M, Soyk, S, Ramakrishnan, S, Wang, X, Goodwin, S, Sedlazeck, FJ, Lippman, ZB, Schatz, MC (2019) Genome Biology doi: https://doi.org/10.1186/s13059-019-1829-6 |
 |
|
| 113. |
The bracteatus pineapple genome and domestication of clonally propagated crops
Chen et al (2019) Nature Genetics doi:10.1038/s41588-019-0506-8 |
 |
|
| 112. |
Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome
Wenger et al (2019) Nature Biotechnology doi:10.1038/s41587-019-0217-9 |
 |
|
| 111. |
Samovar: Single-sample mosaic SNV calling with linked reads
Darby, CA, Fitch, JR, Brennen, PJ, Kelly, BJ, Bir, N, Magrini, V, Leonard, J, Cottrell, CE, Gastier-Foster, JM, Wilson, RK, Mardis, ER, White, P, Langmead, B, Schatz, MC (2019) iScience doi:https://doi.org/10.1016/j.isci.2019.05.037 |
 |
|
| 110. |
Addressing confounding artifacts in reconstruction of gene co-expression networks
Parsana, P, Ruberman, C, Jaffe, AE, Schatz, MC, Battle, A, Leek, JT (2019) Genome Biology https://doi.org/10.1186/s13059-019-1700-9 |
 |
|
| 109. |
Hypo-osmotic-like stress underlies general cellular defects of aneuploidy
Tsai, HJ, Nelliat, AR, Choudhury, MI, Kucharavy, A, Bradford, WD, Cook, ME, Kim, J, Mair, DB, Sun, SX, Schatz, MC, Li R (2019) Nature doi: https://doi.org/10.1038/s41586-019-1187-2 |
 |
|
| 108. |
Duplication of a domestication locus neutralized a cryptic variant that caused a breeding barrier in tomato
Soyk, S, Lemmon, ZH, Sedlazeck, FJ, Jiménez-Gómez, JM, Alonge, M, Hutton, SF, Van Eck, J, Schatz, MC, Lippman, ZB (2019) Nature Plants doi: https://doi.org/10.1038/s41477-019-0422-z |
 |
|
| 107. |
A master regulator of regeneration
Alonge, M, Schatz, MC (2019) Science doi: 10.1126/science.aaw6258 |
 |
|
| 106. |
Clairvoyante: a multi-task convolutional deep neural network for variant calling in Single Molecule Sequencing
Luo, R, Sedlazeck, FJ, Lam, TW, Schatz, MC (2019) Nature Communications doi:https://doi.org/10.1038/s41467-019-09025-z |
 |
|
| 105. |
Graph genomes article collection
Schatz, MC, Cosgrove, A (2019) Genome Biology doi:https://doi.org/10.1186/s13059-019-1636-0 |
 |
|
| 104. |
Novel circular RNA NF1 acts as a molecular sponge, promoting gastric cancer by absorbing miR-16
Wang et al (2019) Endocrine-Related Cancer doi: 10.1530/ERC-18-0478 |
 |
|
| 103. |
Applying Rapid Whole Genome Sequencing to Predict Phenotypic Antimicrobial Susceptibility Testing Results Among Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates
Tamma P.D., Fan Y., Bergman, Y, Pertea, G, Kazmi, A, Lewis, S, Carroll, KC, Schatz, MC, Timp, W, Simner, P.J. (2019) Antimicrobial Agents and Chemotherapy DOI: 10.1128/AAC.01923-18 |
 |
|
| |
| |
2018
|
| 102. |
Genome‐wide patterns of transposon proliferation in an evolutionary young hybrid fish
Dennenmoser, S, Sedlazeck, FJ, Schatz, MC, Altmüller, J, Zytnicki, M, Nolte, AW (2018) Molecular Ecologyhttps://doi.org/10.1111/mec.14969 |
 |
|
| 101. |
Parrot Genomes and the Evolution of Heightened Longevity and Cognition
Wirthlin et al (2018) Current Biologydoi:https://doi.org/10.1016/j.cub.2018.10.050 |
 |
|
| 100. |
Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L.
Zhang et al. (2018) Nature Genetics doi: https://doi.org/10.1038/s41588-018-0237-2 |
 |
|
| 99. |
Conservation genomics of the declining North American bumblebee Bombus terricola reveals inbreeding and selection on immune genes.
Kent, CF, Dey, A, Patel, H, Tsvetkov, N, Tiwari, T, MacPhail, VJ, Gobeil, Y, Harpur, BA, Gurtowski, J, Schatz, MC, Colla, SR, Zayed, A. (2018) Frontiers in Genetics doi: 10.3389/fgene.2018.00316 |
 |
|
| 98. |
KBase: The United States Department of Energy Systems Biology Knowledgebase
Arkin, AP, Steven, RL, Cottingham, RW, Maslov, S, Henry, CS, et al. (2018) Nature Biotechnology doi: 10.1038/nbt.4163 |
 |
|
| 97. |
Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line
Nattestad, M, Goodwin, S, Ng, K, Baslan, T, Sedlazeck, FJ, Rescheneder, P, Garvin, T, Fang, H, Gurtowski, J, Hutton, E, Tseng, E, Chin, C, Beck, T, Sundaravadanam, Y, Kramer, M, Antoniou, E, McPherson, JD, Hicks, J, McCombie, WR, Schatz, MC (2018) Genome Research doi: 10.1101/gr.231100.117 |
 |
|
| 96. |
Precise detection of de novo single nucleotide variants in human genomes
Gomez-Romera, L, Palacios-Flores, K, Reyes, J, Garcia, D, Boege, M, Davila, G, Flores, M, Schatz, MC, Palacios, R (2018) PNAS https://doi.org/10.1073/pnas.1802244115 |
 |
|
| 95. |
Accurate detection of complex structural variations using single molecule sequencing
Sedlazeck, FJ, Reschender, P, Smolka, M, Fang, H, Nattestad, M, von Haeseler, A, Schatz, MC (2018) Nature Methods doi:10.1038/s41592-018-0001-7 |
 |
|
| 94. |
Antibiotic pressure on the acquisition and loss of antibiotic resistance genes in Klebsiella pneumoniae
Simner, PJ, Antar, AA, Hai, S, Gurtowski, J, Tamma, PD, Rock, C, Opene, BNA, Tekle, T, Carroll, KC, Schatz, MC, Timp, W (2018) Journal of Antimicrobial Chemotherapy https://doi.org/10.1093/jac/dky121 |
 |
|
| 93. |
Piercing the dark matter: bioinformatics of long- range sequencing and mapping
Sedlazeck, FJ, Lee, H, Darby, CA, Schatz, MC (2018) Nature Reviews Genetics doi:10.1038/s41576-018-0003-4 [reprint] |
 |
|
| 92. |
Scikit-ribo: Accurate estimation and robust modeling of translation dynamics at codon resolution
Fang, H, Huang, Y, Radhakrishnan, A, Siepel, A, Lyon, GJ, Schatz, MC (2017) Cell Systems https://doi.org/10.1016/j.cels.2017.12.007 |
 |
|
| 91. |
Reference Quality Assembly of the 3.5 Gb genome of Capsicum annuum from a Single Linked-Read Library
Hulse-Kemp, AM, Maheshwari, S, Stoffel, K, Hill, TA, Jaffe, D, Williams, S, Weisenfeld, N, Ramakrishnan, S, Kumar, V, Shah, P, Schatz, MC, Church, DM, Van Deynze, A (2017) Horticulture Research doi:10.1038/s41438-017-0011-0 |
 |
| |
| |
| |
2017
|
| 90. |
LRSim: a Linked Reads Simulator generating insights for better genome partitioning
Luo, R, Sedlazeck, FJ, Darby, CA, Kelly, SM, Schatz, MC (2017) Computational and Structural Biotechnology Journal https://doi.org/10.1016/j.csbj.2017.10.002 |
 |
|
| 89. |
Hybrid assembly with long and short reads improves discovery of gene family expansions
Miller, JR, Zhou, P, Mudge, J, Gurtowski, J, Lee, H, Ramaraj, T, Walenz, BP, Liu, J, Stupar, RM, Denny, R, Song, L, Singh, N, Maron, LR, McCouch, SR, McCombie, WR,
Schatz, MC, Tiffin, P, Young, KD, Silverstein, KAT (2017) BMC Genomics doi: 10.1186/s12864-017-3927-8.
(Please cite this for ECTools hybrid error correction) |
 |
|
| 88. |
16GT: a fast and sensitive variant caller using a 16-genotype probabilistic model
Luo, R, Schatz, MC, Salzberg, SL (2017) GigaScience doi: https://doi.org/10.1093/gigascience/gix045 |
 |
|
| 87. |
Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma
Feigin, ME, Garvin, T, Bailey, P, Waddell, N, Chang, DK, Kelley, DR, Shuai, S, Gallinger, S, McPherson, JD,
Grimmond, SM, Khurana, E, Stein, LD, Biankin, AV, Schatz, MC, Tuveson, DA (2017) Nature Genetics doi:10.1038/ng.3861 |
 |
|
|
| 86. |
GenomeScope: Fast reference-free genome profiling from short reads
Vurture, GW, Sedlazeck, FJ, Nattestad, M, Underwood, CJ, Fang, H, Gurtowski, J, Schatz, MC (2017) Bioinformatics doi: https://doi.org/10.1093/bioinformatics/btx153 |
 |
|
|
| 85. |
Nanopore sequencing meets epigenetics
Schatz, MC (2017) Nature Methods 14, 347–348 (2017) doi:10.1038/nmeth.4240 |
 |
|
|
| 84. |
Bioinformatics of DNA
Heath, LS, Bravo, HC, Caccamo, M, Schatz, MC (2017) Proceedings of the IEEE doi: 10.1109/JPROC.2017.2652958
See entire issue here |
 |
|
|
| 83. |
TGF-β reduces DNA ds-break repair mechanisms to heighten genetic diversity and adaptability of CD44+/CD24- cancer cells
Pal, D, Pertot, A, Shirole, NH, Yao, Z, Anaparthy, N, Garvin, T, Cox, H, Chang, K, Rollins, F, Kendall, J, Edwards, L, Singh, VA,
Stone, GC, Schatz, MC, Hicks, J, Hannon, G, Sordella, R (2017) eLife doi: http://dx.doi.org/10.7554/eLife.21615 |
 |
|
|
2016
|
| 82. |
NanoBLASTer: Fast alignment and characterization of Oxford Nanopore single molecule sequencing reads
Amin, M.R., Skiena, S, Schatz, MC (2016) Proceedings of the Computational Advances in Bio and Medical Sciences (ICCABS) Conference. doi: 10.1109/ICCABS.2016.7802776 |
 |
|
| 81. |
Indel variant analysis of short-read sequencing data with Scalpel
Fang, H, Grabowska, EA, Arora, K, Vacic, V, Zody, MC, Iossifov, I, ORawe, JA, Wu, Y, Jimenez-Barron, LT, Rosenbaum, J, Ronemus, M, Lee, Y, Wang, Z, Lyon, GJ, Wigler, M, Schatz, MC , Narzisi G (2015) Nature Protocols doi:10.1038/nprot.2016.150 |
 |
|
| 80. |
The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation
Lemmon, ZH, Park, SJ, Jiang, K, Van Eck, J, Schatz, MC, Lippman ZB (2016) Genome Research doi: 10.1101/gr.207837.116 |
 |
|
| 79. |
Phased Diploid Genome Assembly with Single Molecule Real-Time Sequencing
Chin, CS, Peluso, P, Sedlazeck, FJ, Nattestad, M, Concepcion, GT, Clum, A, Dunn, C, O'Malley, R, Figueroa-Balderas, R, Morales-Cruz, A, Cramer, GR, Delledonne, M, Luo, C, Ecker, JR, Cantu, D, Rank, DR., Schatz, MC (2016) Nature Methods doi:10.1038/nmeth.4035 |
 |
|
| 78. |
Complete telomere-to-telomere de novo assembly of the Plasmodium falciparum genome through long-read (>11 kb), single molecule, real-time sequencing
Vembar, SS, Seetin, M, Lambert, C, Nattestad, M, Schatz, MC, Baybayan, P, Scherf, A, Smith, ML. (2016) DNA Research doi: 10.1093/dnares/dsw022 |
 |
|
| 77. |
Assemblytics: a web analytics tool for the detection of variants from an assembly
Nattestad, M, Schatz, MC (2016) Bioinformatics doi: 10.1093/bioinformatics/btw369 |
 |
|
| 76. |
Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida
Bombarely, A et al. (2016) Nature Plants doi:10.1038/nplants.2016.74 |
 |
|
| 75. |
Chromosomal-Level Assembly of the Asian Seabass Genome Using Long Sequence Reads and Multi-layered Scaffolding
Vij, S et al. (2016) PLOS Genetics http://dx.doi.org/10.1371/journal.pgen.1005954 |
 |
|
| 74. |
Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius
Rosenfeld, JA et al. (2016) Nature Communication doi:10.1038/ncomms10164 |
 |
|
2015
|
| 73. |
The pineapple genome and the evolution of CAM photosynthesis
Ming, R, VanBuren, R, Man Wai, C, Tang, H, Schatz MC, et al. (2015) Nature Genetics doi:10.1038/ng.3435 |
 |
|
| 72. |
Teaser: Individualized benchmarking and optimization of read mapping results for NGS data
Smolka M, Rescheneder P, Schatz MC, Haeseler AV, Sedlazeck FJ. (2015) Genome Biology 16:235 doi:10.1186/s13059-015-0803-1 |
 |
|
| 71. |
Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome
Goodwin, S, Gurtowski, J, Ethe-Sayers, S, Deshpande, P, Schatz MC*, McCombie, WR* (2015) Genome Research doi: 10.1101/gr.191395.115 |
 |
|
| 70. |
Biological data sciences in genome research
Schatz MC (2015) Genome Research 25: 1417-1422. doi: 10.1101/gr.191684.115 |
 |
|
| 69. |
Metassembler: merging and optimizing de novo genome assemblies
Wences, AH Schatz MC (2015) Genome Biology 16:207. doi:10.1186/s13059-015-0764-4 |
 |
|
| 68. |
Genome and transcriptome of the regeneration-competent flatworm, Macrostomum lignano
Wasik, KA, Gurtowski J, Zhou, X, Mendivil Ramos O, Delás MJ, Battistoni, G, El Demerdash, O, Faciatori, I, Vizoso, DV,
Ladurner, P, Schärer, L, McCombie, WR, Hannon, GJ*, Schatz MC* (2015) PNAS doi: 10.1073/pnas.1516718112 |
 |
|
| 67. |
Interactive analysis and assessment of single-cell copy-number variations.
Garvin, T, Aboukhalil, R, Kendall, J, Baslan, T, Atwal, GS, Hicks, J, Wigler, M, Schatz MC (2015) Nature Methods doi:10.1038/nmeth.3578 |
 |
|
| 66. |
Dual functions of Macpiwi1 in transposon silencing and stem cell maintenance in the flatworm Macrostomum lignano
Zhou X, Battistoni G, Demerdash OE, Gurtowski J, Wunderer J, Falciatori I, Ladurner P, Schatz MC, Hannon GJ, Wasik KA (2015) RNA doi: 10.1261/rna.052456.115 |
 |
|
| 65. |
Big Data: Astronomical or Genomical?
Stephens, Z, et al. (2015) PLOS Biology DOI: 10.1371/journal.pbio.1002195 |
 |
|
| 64. |
Molecular genetic diversity and characterization of conjugation genes in the fish parasite Ichthyophthirius multifiliis
MacColl E, et al. (2015) Molecular Phylogenetics and Evolution 86:1-7. doi:10.1016/j.ympev.2015.02.017 |
 |
|
| 63. |
Extending reference assembly models
Church, DM, et al. (2015) Genome Biology 16:13 doi:10.1186/s13059-015-0587-3 |
 |
|
| 62. |
The challenge of small-scale repeats for indel discovery
Narzisi, G, Schatz MC (2015) Frontiers in Bioengineering and Biotechnology 3:8. doi: 10.3389/fbioe.2015.00008 |
 |
|
|
|
2014
|
| 61. |
Whole genome de novo assemblies of three divergent strains of rice (O. sativa) documents novel gene space of aus and indica
Schatz MC, Maron, LG, Stein, JC, Wences, AH, Gurtowski, J, Biggers, E, Lee, H, Kramer, M, Antoniou, E, Ghiban, E, Wright, MH, Chia, JM, Ware, D, McCouch, S, McCombie, WR (2014) Genome Biology 15:506 doi:10.1186/s13059-014-0506-z |
 |
|
| 60. |
SplitMEM: A graphical algorithm for pan-genome analysis with suffix skips
Marcus, S, Lee, H, Schatz MC (2014) Bioinformatics doi: 10.1093/bioinformatics/btu756 |
 |
|
| 59. |
The contribution of de novo coding mutations to autism spectrum disorder
Iossifov, I et al. (2014) Nature doi:10.1038/nature13908 |
 |
|
| 58. |
Reducing INDEL calling errors in whole-genome and exome sequencing data
Fang, H, Narzisi, G, O'Rawe, J, Wu, Y, Rosenbaum, J, Ronemus, M, Iossifov, I, Schatz MC, Lyon, GJ (2014) Genome Medicine 6:89. doi:10.1186/s13073-014-0089-z |
 |
|
| 57. |
Accurate de novo and transmitted indel detection in exome-capture data using microassembly.
Narzisi, G, O'Rawe, JA, Iossifov, I, Fang, H, Lee, YH, Wang, Z, Wu, Y, Lyon, G, Wigler, M, Schatz MC (2014) Nature Methods doi:10.1038/nmeth.3069 |
 |
|
| 56. |
High-coverage sequencing and annotated assemblies of the budgerigar genome
Ganapathy, G et al. (2014) GigaScience 3:11 doi:10.1186/2047-217X-3-11 |
 |
|
| 55. |
Large-scale sequencing and assembly of cereal genomes using Blacklight
Blood, P, Marcus, S, Schatz MC (2014) XSEDE14 Atlanta GA, July 13-18. 2014 [preprint] |
 |
|
| 54. |
On Algorithmic Complexity of Biomolecular Sequence Assembly Problem
Narzisi, G, Mishra, B, Schatz MC (2014) Algorithms for Computational Biology Lecture Notes in Computer Science, Vol. 8542 [preprint] |
 |
|
2013
|
| 53. |
Cultivation and Complete Genome Sequencing of Gloeobacter kilaueensis sp. nov., from a Lava Cave in Kīlauea Caldera, Hawai'i
Saw JHW, Schatz M, Brown MV, Kunkel DD, Foster JS, et al. (2013) PLoS One 8(10): e76376. doi:10.1371/journal.pone.0076376 |
 |
|
| 52. |
Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species
Bradnam, KR. et al (2013) GigaScience 2:10 doi:10.1186/2047-217X-2-10 |
 |
|
| 51. |
The advantages of SMRT sequencing
Roberts, RJ, Carneiro, MO, and Schatz, MC (2013) Genome Biology 14:405 doi:10.1186/gb-2013-14-6-405 |
 |
|
| 50. |
The DNA60IFX Contest
Schatz, MC, Taylor, J and Schelhorn, S (2013) Genome Biology 14:124 doi:10.1186/gb-2013-14-6-124 |
 |
|
| 49. |
The DNA Data Deluge
Schatz, MC and Langmead, B (2013) IEEE Spectrum July, 2013 |
 |
|
| 48. |
Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.)
Ming, R et al. (2013) Genome Biology 14:R41 |
 |
|
| 47. |
Sixty years of genome biology
Doolittle, WF et al. (2013) Genome Biology 14:113 |
 |
|
| 46. |
Aluminum tolerance in maize is associated with higher MATE1 gene copy number
Maron, LG et al. (2013) PNAS doi: 10.1073/pnas.1220766110 |
 |
|
2012
|
| 45. |
Computational thinking in the era of big data biology
Schatz, MC (2012) Genome Biology 13:177 |
 |
|
| 44. |
Answering the demands of digital genomics
Titmus, MA, Gurtowski, J, Schatz, MC (2012) Concurrency and Computation: Practice and Experience DOI: 10.1002/cpe.2925 |
 |
|
| 43. |
De novo identification of 'heterotigs' towards accurate and in-phase assembly
of complex plant genomes
Price J, et al. (2012) Proceedings of BIOCOMP'12 Las Vegas, NV. [preprint] |
 |
|
| 42. |
Genotyping in the Cloud with Crossbow
Gurtowski, J, Schatz, MC, Langmead, B (2012) Current Protocols in Bioinformatics 39:15.3.1-15.3.15 |
 |
|
| 41. |
The rise of a digital immune system
Schatz, MC, Phillippy, AM (2012) GigaScience 1:4 |
 |
|
| 40. |
Hybrid error correction and de novo assembly of single-molecule sequencing reads
Koren, S, Schatz, MC, Walenz, BP, Martin, J, Howard, JT, Ganapathy, G, Wang, Z, Rasko, DA, McCombie, WR, Jarvis, ED, Phillippy, AM (2012) Nature Biotechnology. doi:10.1038/nbt.2280 |
 |
|
| 39. |
Genomic Dark Matter: The reliability of short read mapping illustrated by the Genome Mappability Score
Lee, H, Schatz, MC (2012) Bioinformatics. 10.1093/bioinformatics/bts330 |
 |
|
| 38. |
Current challenges in de novo plant genome sequencing and assembly
Schatz, MC, Witkowski, J, McCombie, WR (2012) Genome Biology. 12:243 |
 |
|
| 37. |
De novo gene disruptions in children on the autism spectrum
Iossifov, I. et al. (2012) Neuron. 74:2 285-299 |
 |
|
2011
|
| 36. |
Rate of meristem maturation determines inflorescence architecture in tomato
Park, SJ, Jiang, K, Schatz, MC, Lippman, ZB (2011) PNAS. doi: 10.1073/pnas.1114963109 |
 |
|
| 35. |
Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies
Schatz, MC, et al. (2011) Briefings in Bioinformatics. doi: 10.1093/bib/bbr074 |
 |
|
| 34. |
GAGE: A critical evaluation of genome assemblies and assembly algorithms
Salzberg, SL, et al. (2011) Genome Research. doi: 10.1101/gr.131383.111 |
 |
|
| 33. |
Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis.
Donia, MS, et al. (2011) PNAS. doi: 10.1073/pnas.1111712108 |
 |
|
| 32. |
Assemblathon 1: A competitive assessment of de novo short read assembly methods.
Earl, DA, et al. (2011) Genome Research. doi: 10.1101/gr.126599.111 |
 |
|
| 31. |
Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp.
Bogdanove, AJ, et al. (2011) Journal of Bacteriology. doi:10.1128/JB.05262-11 |
 |
|
| 30. |
Rapid Parallel Genome Indexing with MapReduce.
Menon, RK, Bhat, GP, Schatz, MC (2011) Proceedings of the 2nd International Workshop on MapReduce and its applications. HPDC'11, San Jose, CA. [preprint] |
 |
|
2010
|
| 29. |
Quake: quality-aware detection and correction of sequencing reads.
Kelley, DR, Schatz, MC, Salzberg, SL (2010) Genome Biology. 11:R116 |
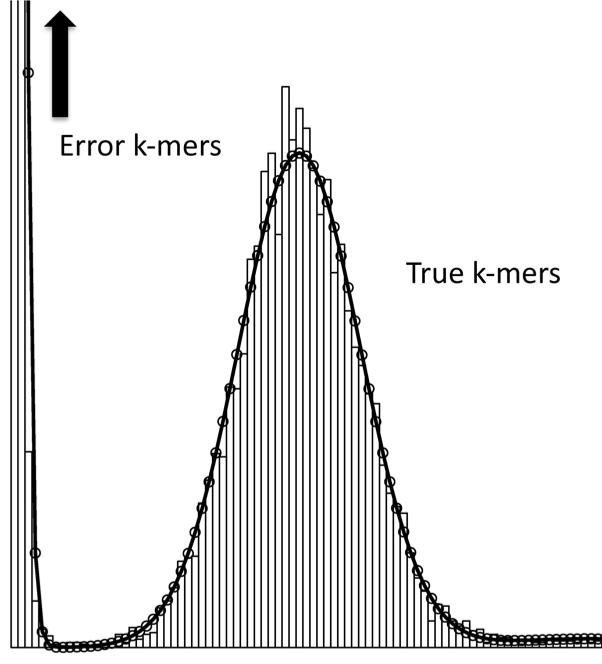 |
|
| 28. |
Genomic survey of the ectoparasitic mite Varroa destructor, a major pest of the honey bee Apis mellifera.
Cornman, SR, Schatz, MC, et al. (2010) BMC Genomics. 11:602 |
 |
|
| 27. |
Multi-Platform Next-Generation Sequencing of the Domestic Turkey (Meleagris gallopavo): Genome Assembly and Analysis.
Dalloul, RA, et al. (2010) PLoS Biology. 8(9): e1000475. |
 |
|
| 26. |
The missing graphical user interface for genomics.
Schatz, MC. (2010) Genome Biology. 11:128 |
 |
|
| 25. |
Design patterns for efficient graph algorithms in MapReduce.
Lin, J., Schatz, MC. (2010) Proceedings of the Eighth Workshop on Mining and Learning with Graphs Workshop (MLG-2010) . |
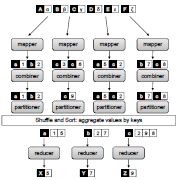 |
|
| 24. |
Cloud Computing and the DNA Data Race.
Schatz, MC, Landmead, B, Salzberg, SL. (2010) Nature Biotechnology. 28:691-693 |
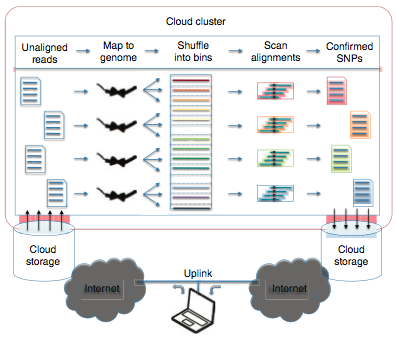 |
|
| 23. |
Integrated microbial survey analysis of prokaryotic communities for the PhyloChip microarray.
Schatz, MC, Phillippy, AM, Gajer, P, DeSantis, TZ, Anderson, GL, Ravel, J. (2010) Applied and Environmental Microbiology. |
 |
|
| 22. |
Assembly of large genomes using second-generation sequencing.
Schatz, MC, Delcher, AL, Salzberg, SL. (2010) Genome Research, 20: 1165-1173. |
 |
|
| 21. |
Assembly complexity of prokaryotic genomes using short reads.
Kingsford, C, Schatz, MC, Pop, M (2010) BMC Bioinformatics 11:21. |
 |
|
2009
|
| 20. |
Searching for SNPs with cloud computing.
Langmead, B, Schatz, MC, Lin, J, Pop, M, Salzberg, SL (2009) Genome Biology 10:R134. |
 |
|
| 19. |
Optimizing data intensive GPGPU computations for DNA sequence alignment.
Trapnell, C, Schatz, MC (2009) Parallel Computing 35(8-9):429-440. |
 |
|
| 18. |
Genomic Analyses of the Microsporidian Nosema ceranae, an Emergent Pathogen of Honey Bees.
Cornman, RS, Chen, YP, Schatz, MC, et al. (2009) PLoS Pathogens 5(6):e1000466. |
 |
|
| 17. |
A whole-genome assembly of the domestic cow, Bos taurus.
Zimin, AV, Delcher, AL, Florea, L, Kelley, DR, Schatz, MC, et al. (2009) Genome Biology 10:R42. |
 |
|
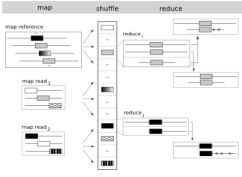
| 16. |
CloudBurst: Highly Sensitive Read Mapping with MapReduce.
Schatz, MC (2009) Bioinformatics 25:1363-1369. |
 |
|
| 15. |
Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps.
Desjardins, CA, et al. (2009) Genome Biology 9:R183. |
 |
|
2008
|
| 14. |
Characterization of Insertion Sites in Rainbow Papaya, the First Commercialized Transgenic Fruit Crop.
Suzuki, JY, et al. (2008) Tropical Plant Biology 1:293-309. |
 |
|
| 13. |
Revealing Biological Modules via Graph Summarization.
Navlakha, S, Schatz, M, Kingsford, C. (2008) Journal of Computational Biology 16(2): 253-264. |
 |
|
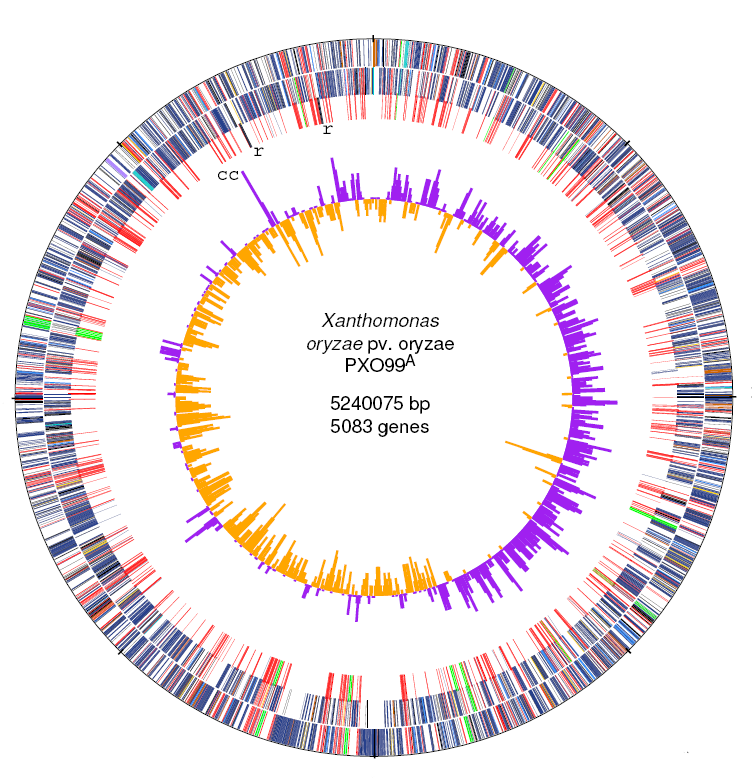
| 12. |
Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A.
Salzberg, SL, Sommer DS, Schatz, MC, et al. (2008) BMC Genomics 9:204. |
 |
|
| 11. |
The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus).
Ming, R, et al. (2008) Nature 452, 991-996. |
 |
|
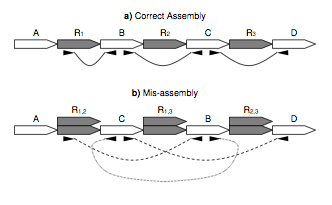
| 10. |
Genome Assembly forensics: finding the elusive mis-assembly.
Phillippy, AM, Schatz, MC, Pop, M. (2008) Genome Biology 9:R55. |
 |
|
2007
|
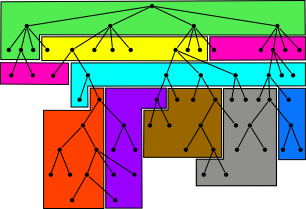
| 9. |
High-throughput sequence alignment using Graphics Processing Units.
Schatz, MC, Trapnell, C, Delcher, AL, Varshney, A. (2007) BMC Bioinformatics 8:474. |
 |
|
| 8. |
Evolution of genes and genomes on the Drosophila phylogeny.
Drosophila 12 Genomes Consortium. (2007) Nature Nov 8;450(7167):203-18. |
 |
|
| 7. |
Draft Genome of the Filarial Nematode Parasite Brugia malayi.
Ghedin, E, et al. (2007) Science 317(5845):1756-1760. |
 |
|
| 6. |
Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus.
Desjardins, CA, et al. (2007) BMC Microbiology 7:61 |
 |
|
| 5. |
Genome Sequence of Aedes aegypti, a Major Arbovirus Vector.
Nene, V et al. (2007) Science 316(5832), 1718-1723. |
 |
|
| 4. |
Hawkeye: a visual analytics tool for genome assemblies.
Schatz, MC, Phillippy, AM, Shneiderman, B, Salzberg, SL. (2007) Genome Biology 8:R34. |
 |
|
| 3. |
Draft Genome Sequence of the Sexually Transmitted Pathogen Trichomonas vaginalis.
Carlton, JM, Hirt, RP, Silva, JC, Delcher, AL, Schatz, M, et al. (2007) Science 315 (5809), 207-212. |
 |
|
2006 & earlier
|
| 2. |
Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species.
Fouts, DE et al. (2005) PLoS Biology 3 (1):e15. |
 |
|
| 1. |
Automated correction of genome sequence errors.
Gajer, P, Schatz, M, Salzberg, SL. (2004) Nucleic Acids Research 32 (2):562-569. |
 |
|
| T6. |
Improving the FAIRness and Sustainability of the NHGRI Resources Ecosystem
Babb et al (2025) arXiv https://arxiv.org/abs/2508.13498 |
 |
|
| T5. |
Modbamtools: Analysis of single-molecule epigenetic data for long-range profiling, heterogeneity, and clustering
Razaghi, R, Hook, PW, Ou, S, Schatz, MC, Hansen, KD, Jain, M, Timp, W (2022) bioRxiv doi: https://doi.org/10.1101/2022.07.07.499188 |
 |
|
| T4 |
Random Forest Factorization Reveals Latent Structure in Single Cell RNA Sequencing Data
Brenerman, BM, Shapiro, MD, Schatz, MC, Battle, A (2021) bioRxiv doi: https://doi.org/10.1101/2021.09.13.460168 |
 |
|
| T3. |
LongTron: Automated Analysis of Long Read Spliced Alignment Accuracy
Wilks, C, Schatz, MC (2020) bioRxiv doi: https://doi.org/10.1101/2020.11.10.376871 |
 |
|
| T2. |
First near complete haplotype phased genome assembly of River buffalo (Bubalus bubalis)
Ananthasayanam, S, Kothandaraman, H, Nayee, N, Saha, S, Baghel, DS, Gopalakrishnan, K, Peddamma, S, Singh, RB Schatz, MC (2019) bioRxiv doi: https://doi.org/10.1101/618785 |
 |
|
| T1. |
SplitThreader: Exploration and analysis of rearrangements in cancer genomes
Nattestad, M, Alford, MC, Sedlazeck, FJ, Schatz, MC (2016) bioRxiv doi: https://doi.org/10.1101/087981 |
 |
|
| 13. |
Algorithms for improving accessibility and representation in large genomic studies
Das, Arun (2025) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 12. |
Methods and applications for large scale pan-genomic analysis
Jenike, Katharine (2024) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 11. |
Computational approaches for discovery of gene regulatory mechanisms
Starostik, Margaret (2024) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 10. |
Genomic Variant Calling at Scale Across Human Populations
Zarate, Samantha (2023) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 9. |
Computational Methods for Structural Variation Analysis in Populations
Kirsche, Melanie (2022) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 8. |
Computational Methods for Genomics, Transcriptomic, and Epi-Omic Analysis with Long-Read Sequencing
Kovaka, Sam (2022) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 7. |
Pan-Genomics and the Structural Diversity of Plant Genomes
Alonge, Michael (2021) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 6. |
Mathematical Analysis of Genome Complexity and Population Diversity using Next- and Third-Generation Sequencing Technologies
Ranallo-Benavidez, Timothy (2020) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 5. |
Computational Methods Addressing Genetic Variation in Next-Generation Sequencing Data
Darby, Charlotte (2020) Ph.D. Disseration, Johns Hopkins University |
 |
|
| 4. |
Graphical and machine learning algorithms for large-scale genomics data
Fang, Han (2017) Ph.D. Disseration, Stony Brook University |
 |
|
| 3. |
Computational methods for analysis and visualization of long-read sequencing data in cancer genomics
Nattestad, Maria (2017) Ph.D. Disseration, Cold Spring Harbor Laboratory |
 |
|
| 2. |
Algorithms to Aid Cancer Genomics with Advanced Sequencing Technology.
Garvin, Tyler (2016) Ph.D. Disseration, Cold Spring Harbor Laboratory |
 |
|
| 1. |
Algorithms and Applications in Genome Assembly using Long Read Sequencing Technology.
Lee, Hayan (2015) Ph.D. Disseration, Stony Brook University |
 |
|
| 0. |
High Performance Computing for DNA Sequence Alignment and Assembly.
Schatz, M.C. (2010) Ph.D. Disseration, Department of Computer Science, University of Maryland |
 |
|
See more in Google Scholar.
|
|
